PCA: Principal Component Analysis
2026-03-10
Plan
What is PCA and why do we use it?
Demonstration with gapminder data in 3 dimensions
More demonstrations
What is PCA?
A tool for simplifying data sets with many variables down to a small number
Works best with quantitative variables that are partially correlated
Can be used with one or more categorical variables to identify subgroups
-
Main results are
- Spatial pattern of points (with categorical labels)
- Directions of principal components along original axes (loadings)
- Amount of variation along each PC axis
Be careful with scaling and units
Example data
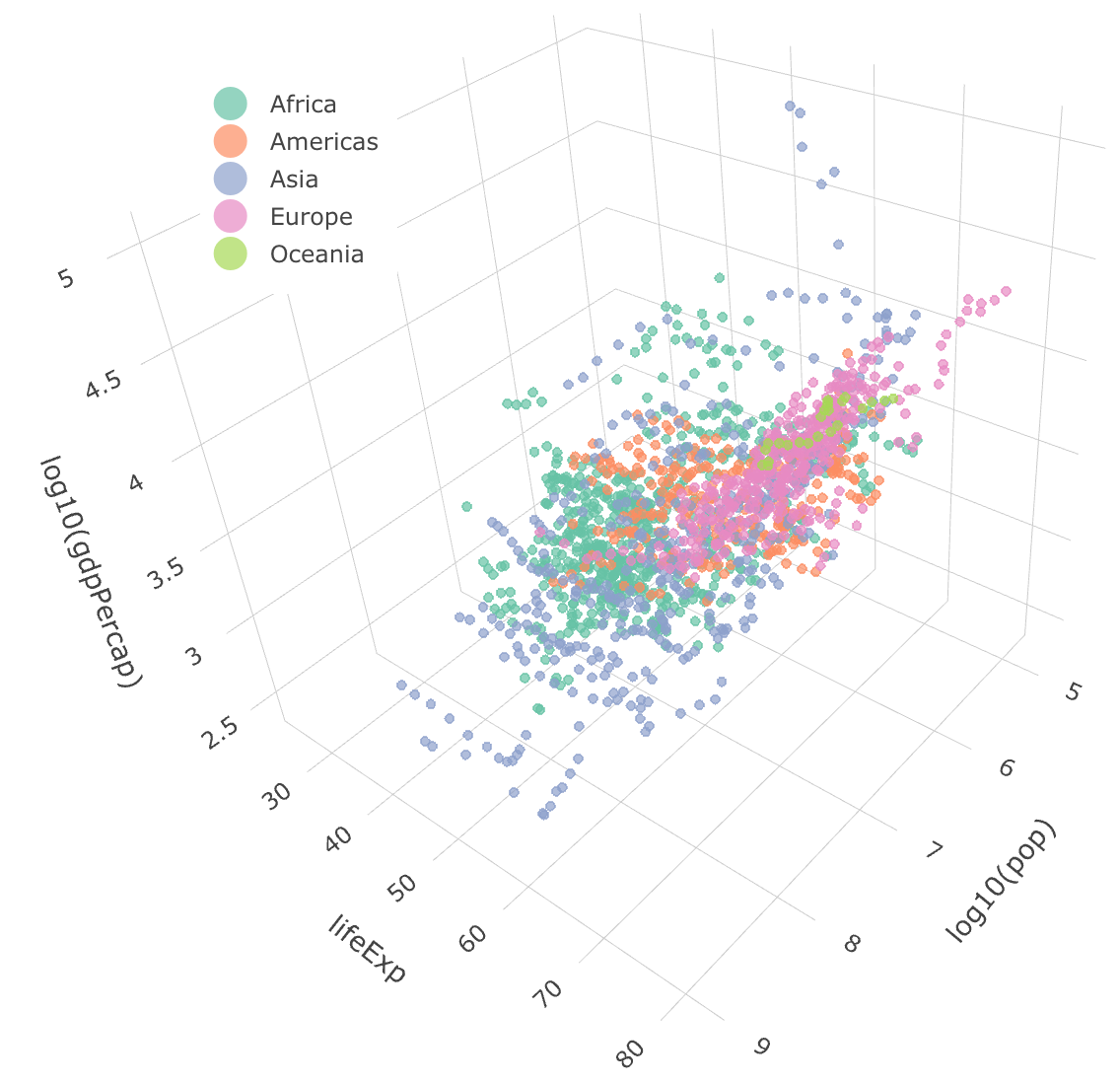
Plot made with plot_ly function in plotly package.
Pairs plot
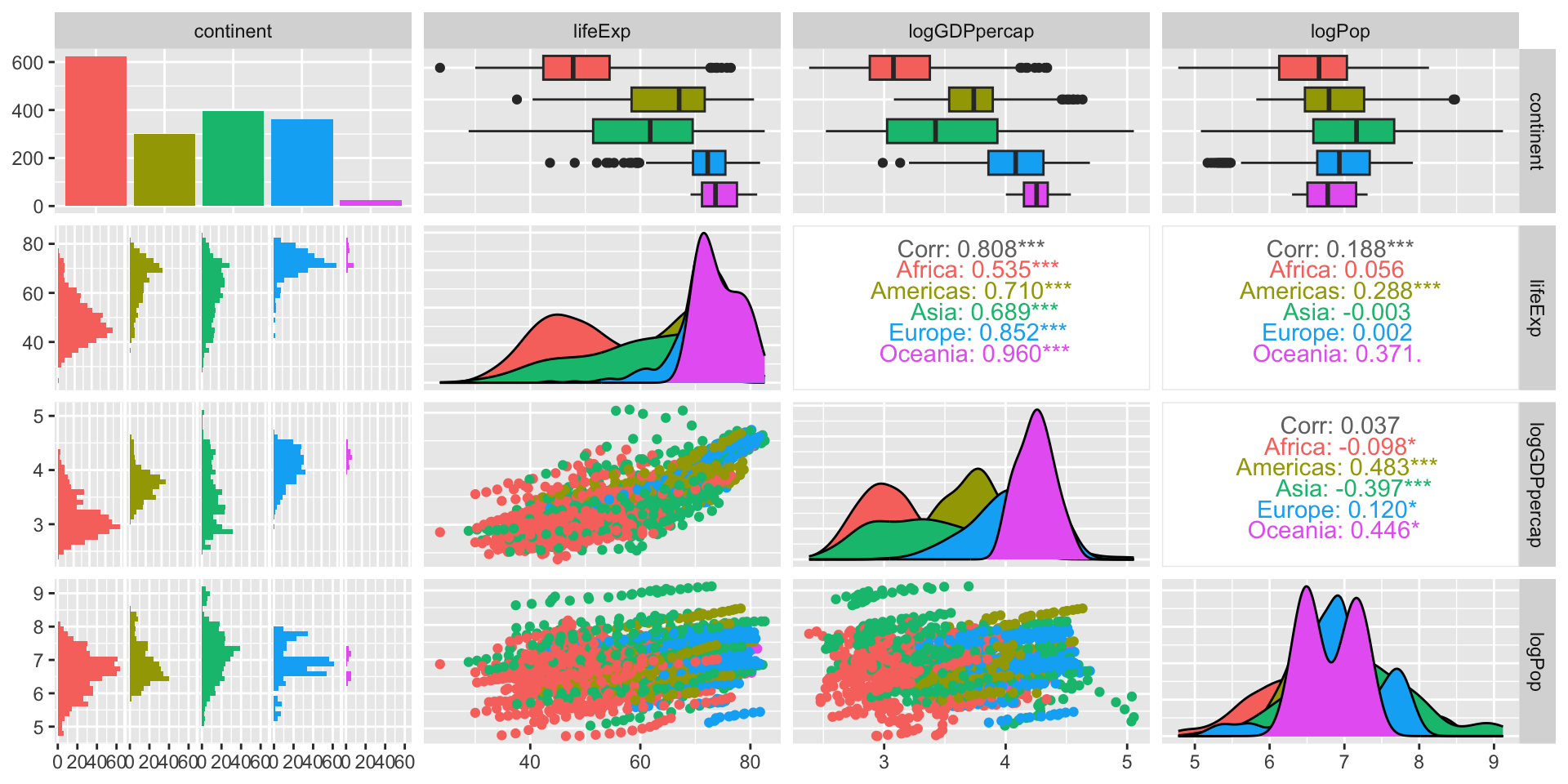
ggpairs is in the GGally package.
PCA: 2007 data only
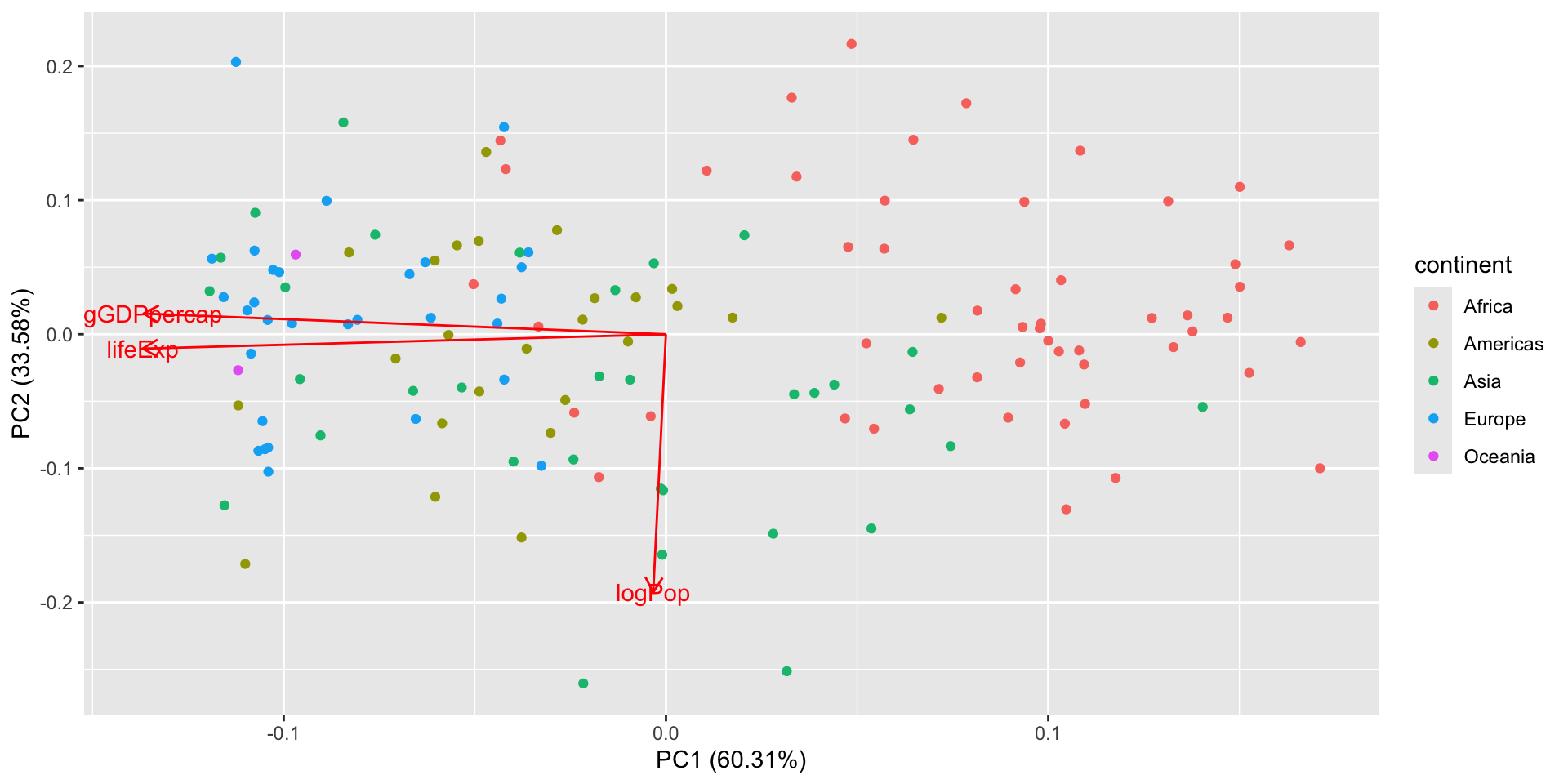
Loadings
This matrix tells you how to rotate the data into PC coordinates which is the weight of each original variable in the computation of PCs.
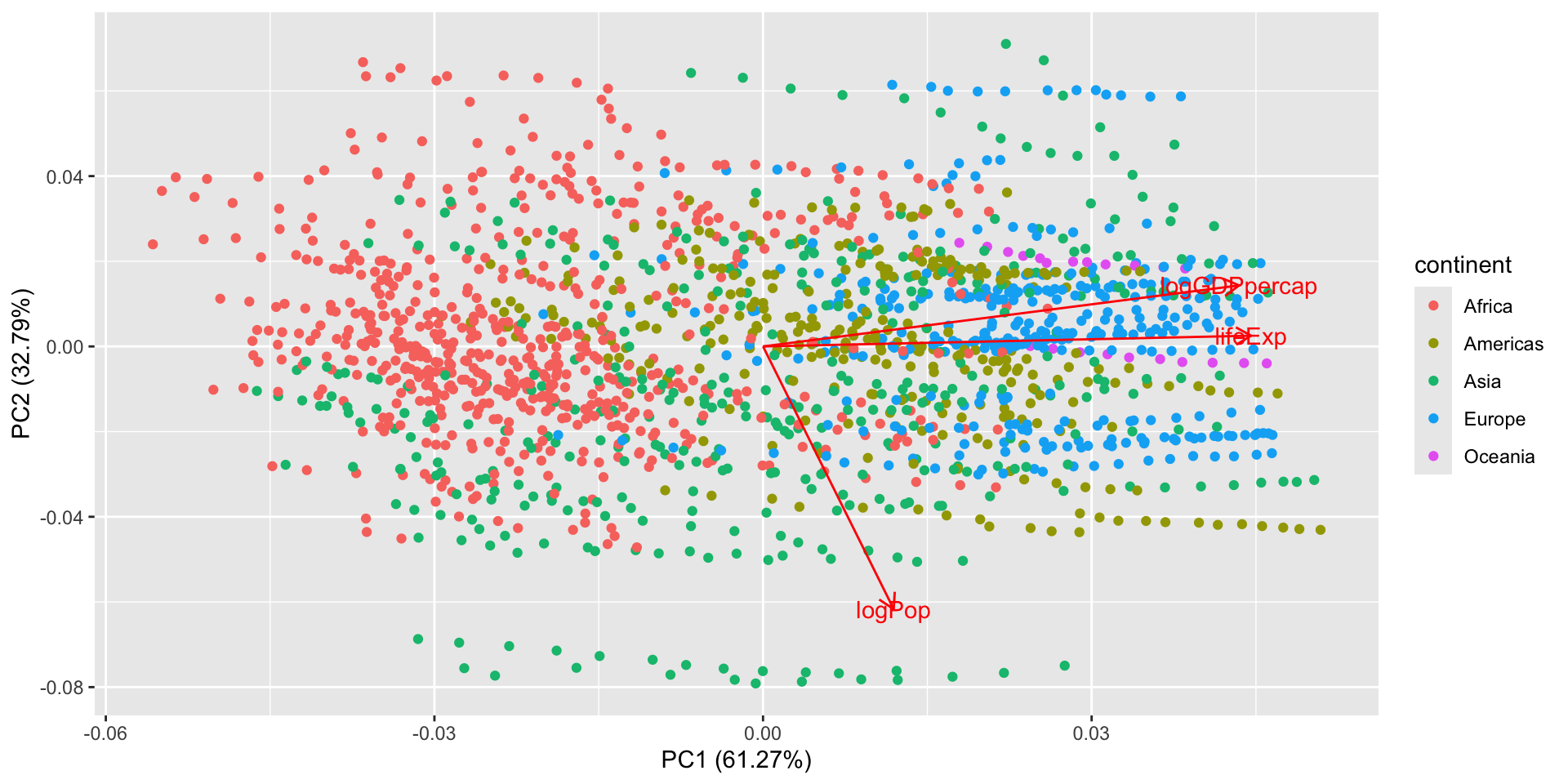
PCA: All years
Penguins
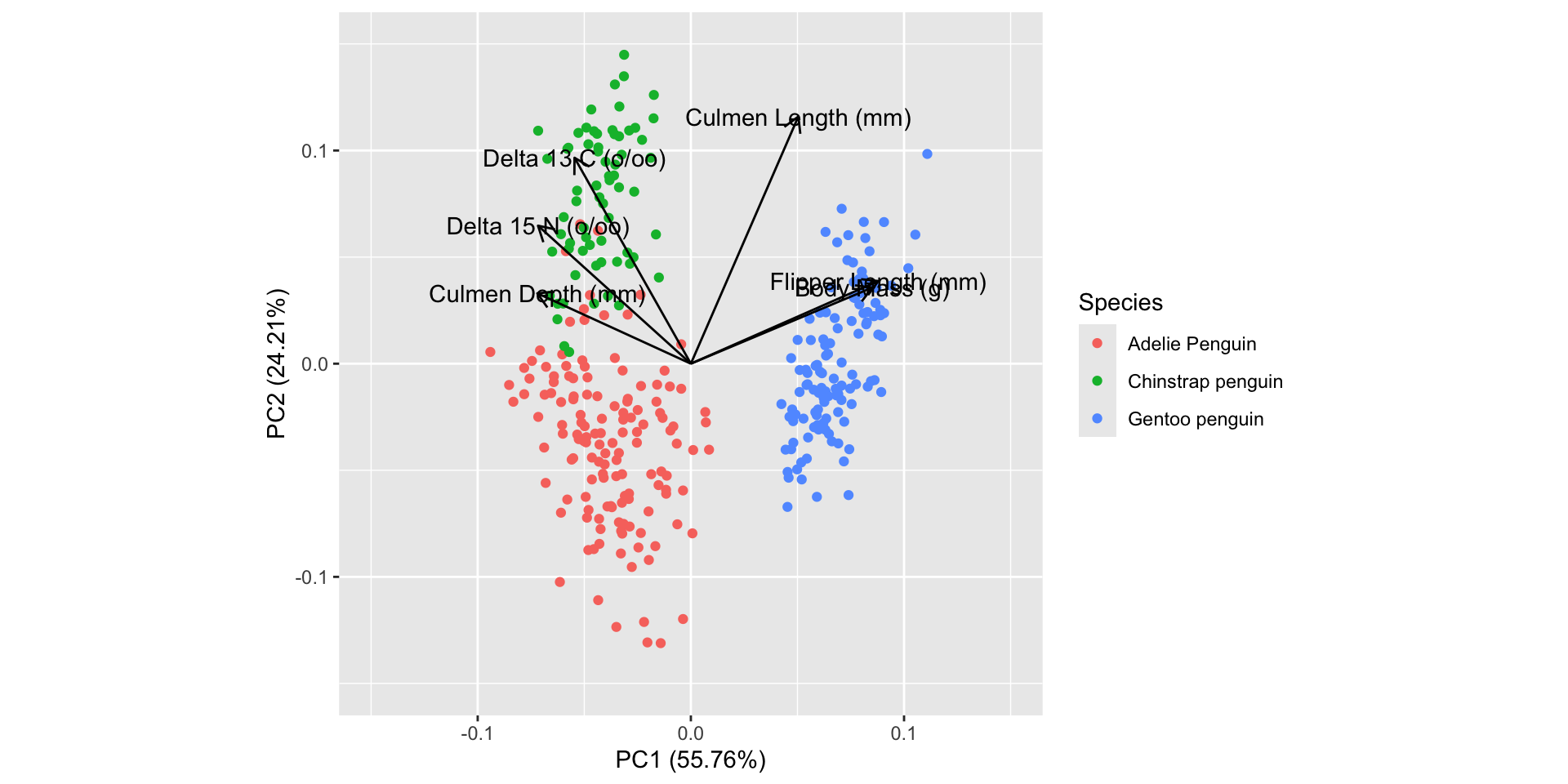
pca2 %>% tidy(matrix="loadings") |> # same as rotation
filter(PC < 3) |>
pivot_wider(values_from="value",
names_from="PC", names_prefix="PC_") |> kable() |> kable_styling(full_width = FALSE)| column | PC_1 | PC_2 |
|---|---|---|
| Culmen Length (mm) | 0.2871721 | 0.6602934 |
| Culmen Depth (mm) | -0.4102740 | 0.1879624 |
| Flipper Length (mm) | 0.5008652 | 0.2207240 |
| Body Mass (g) | 0.4846484 | 0.2033560 |
| Delta 15 N (o/oo) | -0.4084370 | 0.3686830 |
| Delta 13 C (o/oo) | -0.3108644 | 0.5501663 |
Summary
Use PCA to reduce the number of dimensions (variables) in your data
Interpret loadings (arrows and numeric vectors)
Pay attention to the proportion of variance along each principal component
Consequences of scaling variables (or not scaling them)
Exercises
-
Practice the PCA skills in this lesson
- Both writing and fixing R code for a new dataset, and
- Interpreting and getting insight from the results
Penguin PCA code
See the course notes for more code examples.
my_penguins_raw = penguins_raw |> select(-`Sample Number`) |>
select(Species, where(is.numeric) ) |> na.omit() |>
mutate(Species = str_remove(Species, "\\(.*\\)"))
pca2 <- my_penguins_raw |> select(-Species) |> prcomp(scale=TRUE)
autoplot(pca2, data = my_penguins_raw,
loadings=TRUE, loadings.label = TRUE,
loadings.label.colour = "black", loadings.colour = "black",
colour = 'Species') + xlim(-0.15, 0.15) + ylim(-0.15, 0.15)